AI-Powered Innovative R&D Platform for Botanical Drugs
This platform represents a dedicated effort by our AI R&D team to address critical bottlenecks in botanical drug development—including prolonged R&D timelines, unclear mechanisms of action, complex active ingredient profiles, and fragmented knowledge. We are researching and building a specialized AI innovation platform that integrates large language models (LLMs), Retrieval-Augmented Generation (RAG), and fine-tuning techniques.
By systematically consolidating data from classical herbal texts, modern scientific literature, patents, and proprietary experimental datasets—and further integrating cellular nutrition and bio-fermentation technologies—we are constructing a comprehensive, botanical-drug-specific knowledge graph. This graph spans the entire R&D chain, linking “herbs → chemical constituents → molecular targets → biological pathways → syndromes (TCM patterns) → formulations.”
Technically, the platform adopts a tripartite architecture: “Large Model + RAG + LoRA Fine-tuning.” It uses advanced LLMs such as DeepSeek or Qwen as the foundation. RAG, enhanced with vector feedback mechanisms, dynamically injects up-to-date domain knowledge to improve response accuracy. Additionally, the model undergoes LoRA-based fine-tuning using expert-annotated instruction datasets, enabling it to master complex reasoning paradigms unique to botanical drugs—particularly the synergistic effects of multi-component formulations.
Initial validation has already demonstrated the feasibility of this technical approach. The platform holds strong promise to reduce the traditional botanical drug R&D cycle from 4–6 years to just 1–2 years, significantly lowering costs while enhancing overall R&D efficiency.
RAG+Vector FeedBack Inference Engine
The Engine Layer is a high-performance service built based on FastAPI. Its core workflow, as shown in Figure, includes the following key steps:
Step 1: LLM-Based Syndrome Differentiation Analysis. After receiving the user's symptom description, the system first utilizes the DeepSeek-Chat LLM for TCM syndrome differentiation to identify 1-3 most probable syndromes (Zheng Hou). This step simulates the thinking of a TCM practitioner and lays the foundation for accurate subsequent recommendations.
Step 2: Syndrome Knowledge Retrieval. The identified syndromes are re-ranked to pinpoint the main syndrome, which is then encoded into a vector. Relevant TCM theoretical knowledge is retrieved from the syndrome knowledge base, including clinical manifestations, treatment principles, commonly used formulas and herbs for that syndrome, etc.
Step 3: Feedback-Enhanced Context Construction. The system uses BGE-M3 to encode the current symptoms and performs vector similarity search in the PostgreSQL feedback database. It aggregates recommended items that received high scores (e.g., ≥8.0) under semantically similar symptoms as positive preferences and also identifies those with universally low scores (e.g., ≤3.0) as negative warnings, together constituting the "User Feedback Reference" context.
Step 4: LLM Inference and Generation. The syndrome knowledge summary and the feedback-enhanced context are injected into the prompt for the DeepSeek-Chat LLM. Guided by these structured inputs and adhering to preset TCM principles (e.g., prioritizing classics, safety first), the LLM finally outputs a natural language response containing syndrome differentiation analysis, reasoning for the recommendation, and detailed information on specific formulas and herbs. This process strictly constrains the LLM's generation boundaries, effectively mitigating its inherent "hallucination" problem.
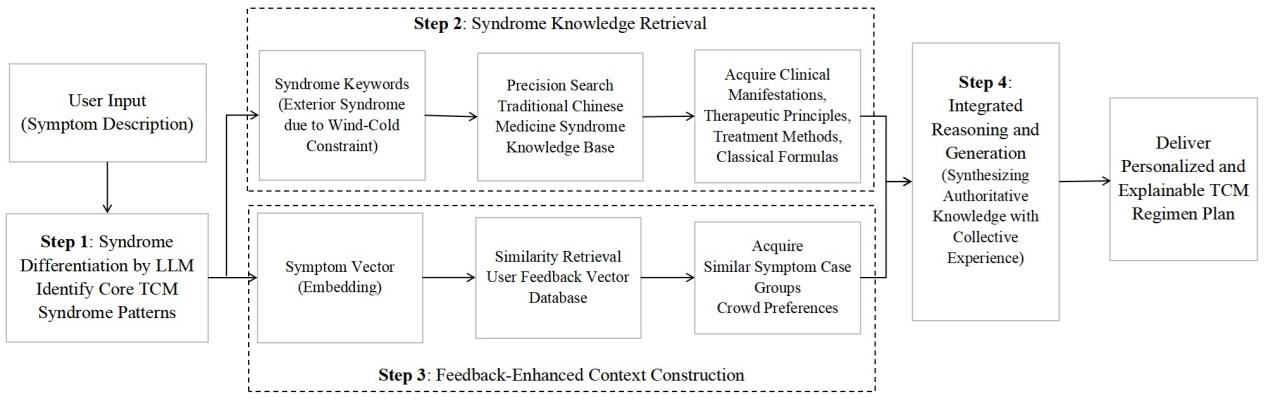
Core workflow of the engine layer
Solution Architecture
Comprehensive Coverage: Complete chain from bottlenecks → technologies → data → knowledge → AI → applications → value
Trinity Architecture: Synergistic integration of Large Models + RAG + LoRA Fine-Tuning
Knowledge Panorama: End-to-end knowledge graph covering "Medicinal Materials - Components - Targets - Pathways - Symptom Patterns - Formulations"
Technological Fusion: Deep integration of AI with cell nutrition, bioprocessing, and other biotechnologies
Data-Driven Approach: Multi-source fusion of ancient texts, modern literature, patent data, and experimental data
Quantifiable Value: Clear metrics for cycle reduction, cost savings, and efficiency improvements
Feedback Loop: Continuous model optimization driven by experimental validation feedback

Solution Architecture
